How complex is epigenetics?
Author: Victor Solis; September 10, 2019
At a molecular level, the expression of a gene can be regulated during its transcription to RNA or during its translation to proteins. A gene that is prevented from generating proteins, cannot exert its function within a cell or express its phenotypic trait on an organism. As a result a wide range of determinant biological processes are impacted, including normal development or disease progression.
Epigenetics engloves at least 3 different mechanisms that regulate the expression of genes. Epigenetics exerts regulation via covalent modifications (DNA methylation and histone modifications), transcription factors and microRNAs.
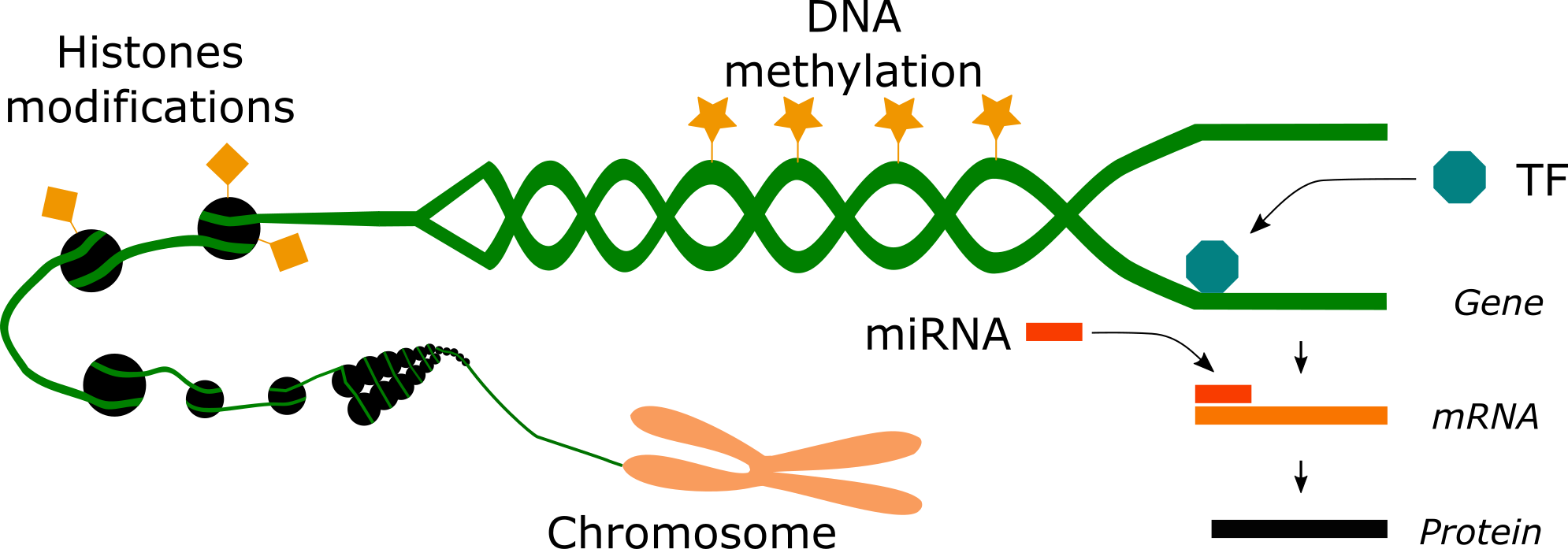
As explained in the previous article, covalent modifications include DNA methylation and hydroxymethylation on cytosine residues at specific regions of the genome called CpG sites. They also include histone modifications such as lysine and arginine methylation and acetylation as well as serine and threonine phosphorylation and lysine ubiquitination and sumoylation. Covalent modifications exert control over the genome by (1) remodeling the structure of the chromatin, making genes either less or more accesible for expression and/or by (2) recruiting or preventing the binding of proteins required for the expression of the genome. The mechanism of histone modifications is more ambiguous than DNA methylation. While DNA methylation works often as a suppresive or silencing factor of gene expression, histone modifications are likely to function in different modes. An acetylated histone at one genome position may exert opposite effects than an acetylated histone at another position of the genome. Similarly, a methylation at one histone lysine can have a different effect than the methylation of another lysine on the same histone. Covalent modifications are imprinted by proteins called writers and are cleaned out by proteins called erasers. Drugs that inhibit writers or erasers are nowadays used for medical treatments of certain kinds of cancer. Other histone modifications associated with other diseases and cancers await to be discovered.
Transcription factors (TF) are another mechanism of epigenetic regulation. They can act as positive or negative regulators of the expression of genes. Transcription factors bind to DNA sequences that are located at specific regions of the genome. These sequences can be either enhancers of transcription or insulators of a negative or positive effect on transcription. Enhancers serve as hubs for transcription factors that promote the expression of a gene. Insulators can either stop the supressive effect of DNA methylation or the promotive effect of enhancers.
MicroRNAs (miRNAs) are RNA molecules of 17-25 nucleotides that do not encode any protein and thus are never translated. However, they exert suppresive roles on the expression of a number of genes. It has been calculated that each miRNA can downregulate the translation of about 100 to 200 messenger RNAs (mRNAs: RNAs that encode for proteins). MicroRNAs function via pairing with mRNAs, which results in the degradation of the mRNA for most cases or in the downregulation of their translation into proteins in some other cases.
At EpiQMAx, we have experts that focus on a subset of the epigenetic world: histone modifications. Together with the pharmaceutical industry, clinics and other partners, we want to decode the unknown associations of these modifications with diseases in humans.